Project
MAT4COVID: Interaction of SARS-CoV-2 virus with materials: a multi computational simulation study.
Marie Sklodowska-Curie Actions Individual Fellowship (MSCA-IF Grant agreement ID: 101026158)
Principal investigator: Dr. Jordi Faraudo;
Experienced researcher: Dr. Mehdi Sahihi;
Abstract
Importance
Epidemic outbreaks of respiratory viral diseases represent a serious issue to public health, as demonstrated historically by influenza pandemics[1] and now by the ongoing COVID-19 pandemic. The SARS-CoV-2 virus (responsible for the COVID-19 disease) is the third documented spill over of an animal coronavirus to humans in only two decades[2] and it has the highest transmission rate among them[3]. The outbreak originated in December 2019 in Wuhan, China and expanded so fast around the world that the World Health Organization’s (WHO) declared a Public Health Emergency of International Concern (PHEIC) on 30 January 2020 followed by a declaration of global pandemic on 11 March 2020,[4],[5]. As of February 20, 2022, over 424 million people were infected. Breaking the chain of transmission is now a top priority to control this disease[6]. Transmission is due to respiratory secretions or droplets expelled by infected individuals. These secretions may affect other persons (direct transmission) but also they are able to contaminate surfaces. Viable SARS-CoV-2 virus can remain viable on surfaces for periods ranging from hours to days, depending on the ambient environment (including temperature and humidity) and the type of surface[7]. Transmission also occur indirectly through touching surfaces, textiles or objects contaminated with virus followed by touching the mouth, nose, or eyes. For this reason, generic antiviral disinfection measures (appropriate for previously known enveloped viruses) are recommended for hands, surfaces and materials, e.g. the use of disinfectants containing alcohol or soaps with surfactants. Design of more focused recommendations, more efficient disinfestation strategies and development of virucidal surfaces or textiles will be possible with a fundamental knowledge of the physico-chemical aspects of the interaction of the virus with materials. Experimental evidence[8] suggests that the interaction of the virus with surfaces of different materials is highly specific but at the present time, the fundamental aspects of the virus-material interactions are not known.
The objective of this project is to study the fundamental physico-chemical aspects of the virus-surface interaction in order to identify which factors (hydrophobic or hydrophilic nature of the surface, surface charge, nanostructuration or others) make a surface prone to virus adhesion or, on the contrary, make a surface virucidal.
[1] US Centers for disease control and prevention (Past Pandemics summary) +info
[2] Zhou P, Yang XL, Wang XG, et al., Nature, 2020, (579), 270–273.
[3] MacIntyre CR., Global Biosecurity, 2020, (1), 1–3.
[4] Chan JFW, Yuan S, Kok KH, et al., Lancet, 2020, (395), 514–523.
[5] Chen N, Zhou M, Dong X, et al. Lancet, 2020, (395), 507–513.
[6] Transmission of SARS-CoV-2 – implications for infection prevention precautions: Scientific brief, WHO, 09 July 2020.
[7] Van Doremalen N, Bushmaker T, Morris DH, et al., N Engl J Med, 2020, (382), 1564-1567.
[8] Kampf G, Todt D, Pfaender S, Steinmann E, J Hosp Infect, 2020, (104), 246–251.
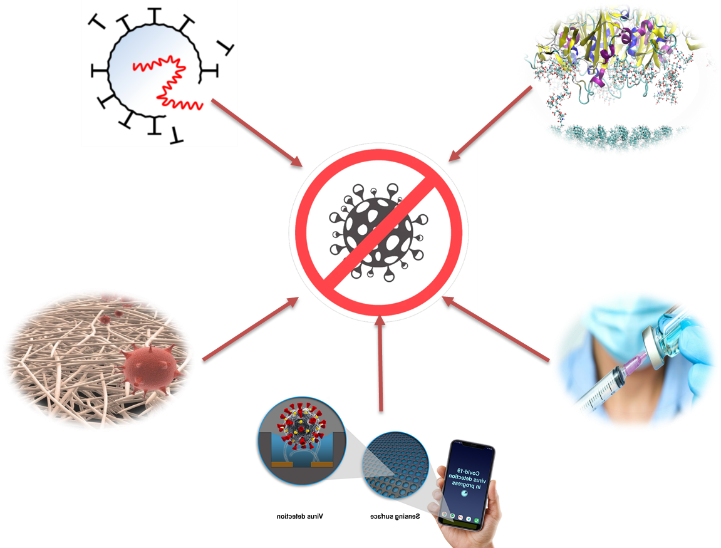
Figure 1: Key steps where the MAT4COVID project could counter the disease.
As a multidisciplinary perspective encompassing diverse fields such as virology, biology, medicine, engineering, chemistry, materials science, and computational science, our project can support the fight against COVID-19, as well as infectious diseases in general, including future pandemics.
Considering what we know so far about the life cycle of the virus, we envision key steps where the MAT4COVID project could counter the disease (Figure 1):
- Virus inactivation
- To understand the mechanism of interactions
- Vaccine design
- To design a virus removal filter
- Virus detection
Structure of the SARS-CoV-2 virus
The structure of the SARS-CoV-2 virus is well known: it has the typical structure of a coronavirus with an envelope containing lipids and proteins (Figure 2), which protects the nucleocapsid that packages the viral RNA. The large protruding glycoprotein spikes (S) on the envelope, typical of the Coronaviridae family of viruses, are responsible for the interaction with host cell receptors and with the environment. The molecular structure with atomistic coordinates of the SARS-CoV-2 virus spike was published[9] as early as in March 2020.

Figure 2. (a) Electron microscopy image of a SARS-CoV-2 coronavirus particle, freely distributed by the NIAID’s Rocky Mountain Laboratories (NIAID-RML), colored to emphasize the virus structure. The spikes protruding from the virus envelope (in yellow color) are clearly visible. Typical diameter ranges from 80 nm to 120 nm. (b) General scheme of a coronavirus indicating their main structural features. We show the nucleocapsid (purple) that packages the viral RNA and the viral envelope made by lipids (pink), envelope protein E (blue), membrane protein M (red) and the protruding spike glycoproteins (green). The scheme was made using CellPAINT software (c) Visualization of an atomistic model of the SARS-CoV-2 spike glycoprotein in the open state inserted into a fully hydrated lipid bilayer (water and ions not shown). The atomic coordinates (a total of 1.7×106 atoms) were downloaded from posted data at the COVID-19 repository12 and represented using Visual Molecular Dynamics software. The glycans that cover the S protein are shown in orange in Van der Waals size. (d) S protein from (c) colored to indicate its main regions. The S protein is synthesized as a single 1273 amino acid polypeptide chain, which associates as a trimer. Each trimer can be divided into the head, stalk, and cytoplasmic domain (CP) or cytoplasmic tail (CT), indicated in the figure. As the S protein is activated for infection, it releases the fusion peptide (FP) indicated in the figure, which penetrates and primes the host cell membrane for fusion. The most external part contains an N-terminal domain (NTD) and the receptor binding domain (RBD), responsible for the interaction with the angiotensin-converting enzyme 2 (ACE2) receptor to gain entry into the host. The different parts are connected by the central helix (CH), and the connecting domain (CD). Domains that are not resolved via cryo-EM or X-ray experiments include the heptad repeat 2 (HR2) and the transmembrane (TM) domains forming the stalk, and the CT.
[9] Wrapp D, Wang N, Corbett KS, et al. Science, 2020, (367), 1260-1263.
Issues Concerning Survival of Viruses on Surfaces




